A wrinkle in timers: evolutionary rewiring of conserved biological timekeepers.
Spangler R, Jonnalagadda K, Ward J, Partch CL
(2025)
Trends in Biochemical Sciences 50: 344-55

PERspectives on circadian cell biology.
Mihut A, O'Neill JS, Partch CL, Crosby P
(2025)
Philos Trans R Soc Lond B Biol Sci doi: 10.1098/rstb.2023.0483

Markovian State Models uncover Casein Kinase 1 dynamics that govern circadian period.
Ricci CG, Philpott JM, Torgrimson MR, Freeberg AM, Narasimamurthy R, de Barros EP, Amaro R, Virshup DM, McCammon JA, Partch CL
(2025)
bioRxiv https://www.biorxiv.org/content/10.1101/2025.01.17.633651v1

Potassium rhythms couple the circadian clock to the cell cycle.
Rodríguez SG, Crosby P, Hansen LL, Grünewald E, Beale AD, Spangler RK, Rabbitts BM, Partch CL, Stangherlin A, O'Neill JS, van Ooijen G
bioRxiv https://www.biorxiv.org/content/10.1101/2024.04.02.587153v1

Temperature-Dependent Fold-Switching Mechanism of the Circadian Clock Protein KaiB.
Zhang N, Sood D, Guo SC, Chen N, Antoszewski A, Marianchuk T, Chavan A, Dey S, Xiao Y, Hong L, Peng X, Baxa M, Partch C, Wang LP, Sosnick TR, Dinner AR, LiWang A
(2024)
Proc Natl Acad Sci USA doi: 10.1073/pnas.2412327121
bioRxiv https://www.biorxiv.org/content/10.1101/2024.05.21.594594v1

A conserved chronobiological complex times C. elegans development.
Spangler RK*, Ashley GE*, Braun K*, Wruck D, Ramos-Coronado A, Ragle JM, Iesmantavicius V, Hess D, Partch CL, Großhans H, Ward JD
bioRxiv https://www.biorxiv.org/content/10.1101/2024.05.09.593322v1


CHRONO participates in multi-modal repression of circadian transcriptional complexes.
Crosby P, Goularte NF, Sharma D, Chen E, Parico GCG, Philpott JM, Harold R, Gustafson CL, Partch CL
bioRxiv https://www.biorxiv.org/content/10.1101/2022.10.04.510902v1

Isoform-specific C-terminal phosphorylation drives autoinhibition of Casein Kinase 1.
Harold R*, Tulsian NK*, Narasimamurthy R, Yaitanes N, Ayala Hernandez MG, Lee H-W, Virshup DM, Partch CL
(2024)
Proc Natl Acad Sci USA doi: 10.1073/pnas.2415567121
bioRxiv https://www.biorxiv.org/content/10.1101/2023.04.24.538174v1

AI is a viable alternative to high throughput screening: a 318-target study
Atomwise AIMS Program
Scientific Reports
(2024)
Apr 2;14(1):7526. doi: 10.1038/s41598-024-54655-z

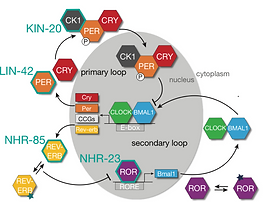
Regulation of the circadian clock in C. elegans by clock gene homologs kin-20 and lin-42.
Lamberti ML, Spangler RK, Cerdeira V, Ares M, Rivollet L, Ashley GE, Ramos Coronado A, Tripathi S, Spiousas I, Ward JD, Partch CL, Bénard CY, Goya ME, Golombek DA
Scientific Reports
(2024)
Jun 5;14(1):12936. doi: 10.1038/s41598-024-62303-9
PDB coordinates for C. elegans LIN-42 PAS-B domain: 8GCI



Conformationally responsive dyes enable protein-adaptive differential scanning fluorimetry.
Wu T, Yu JC, Suresh A, Gale-Day ZJ, Alteen MG, Woo AS, Millbern Z, Johnson OT, Carroll EC, Partch CL, Fourches D, Vinueza NC, Vocadlo DJ, Gestwicki JE
Nature Biotechnology
(2024)
May 14. doi: 10.1038/s41587-024-02158-7


Dimerization Rules of Mammalian PAS Proteins.
Rojas BL, Vazquez-Rivera E, Partch CL, Bradfield CA
J Mol Biol

PAS Dimerization at the Nexus of the Mammalian Circadian Clock.
Sharma D and Partch CL
J Mol Biol

The inner workings of an ancient biological clock.
Fang M, LiWang A, Golden SS, Partch CL
Trends in Biochemical Sciences



Cooperation between bHLH transcription factors and histones for DNA access.
Michael A*, Stoos L*, Crosby P, Eggers N, Nie XY, Makasheva K, Minnich M, Healy KH, Weiss J, Kempf G, Cavadini S, Kater L, Seebacher J, Vecchia L, Chakraborty D, Isbel L, Grand RS, Andersch F, Fribourgh JL, Schübeler D, Zuber J, Liu AC, Becker PB, Fierz B, Partch CL, Menet J, Thomä N
Nature
(2023) 619: 385-393
Featured in Murawska M, Ladumer, Margulies CE (2023) Pioneers conquer core histones at the chromatin frontier. Nat Struct Mol Biol 30: 1050-1053
PDB coordinates for CLOCK:BMAL1-nuc SHL+5.8 – 8OSK
PDB coordinates for CLOCK:BMAL1-nuc SHL-6.2 – 8OSJ
PDB coordinates for CLOCK:BMAL1-nuc native Por promoter – 8OSL
PDB coordinates for OCT4 and MYC:MAX-nuc – 8OTS
(EMD 17183)
PDB coordinates for MYC:MAX-nuc SHL+5.8 – 8OTT
(EMD 17184)
ChIP-Seq data of MYC-MAX and CLOCK-BMAL1 on in vitro reconstituted chromatin, GEO accession code GSE224589

Protocols for in vitro reconstitution of the cyanobacterial circadian clock.
Chavan A, Heisler J, Chang Y-G, Golden SS, Partch CL, LiWang A
Biopolymers

PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Philpott JM, Freeberg AM, Park J, Lee K, Ricci CG, Hunt SR, Narasimamurthy R, Segal DH, Robles R, Cai YD, Tripathi S, McCammon JA, Virshup DM, Chiu JC, Lee C, Partch CL
(bioRxiv https://www.biorxiv.org/content/10.1101/2022.06.24.497549v1)
Meet the Authors: Jonathan Philpott and Carrie L. Partch
PDB coordinates for CK1-human PER2 2p-FASP complex: 8D7M
PDB coordinates for CK1-human PER2 3p-FASP complex: 8D7N
PDB coordinates for CK1-human PER2 4p-FASP complex: 8D7O
PDB coordinates for CK1-Drosophila PER pShort complex: 8D7P
Chemical shift assignments for the human PER2 FASP region at BMRB

How circadian clocks keep time––the discovery of slowness.
Partch CL and Brunner M
FEBS Letts

Coupling of distant ATPase domains in the circadian clock protein KaiC.
Swan JA*, Sandate CR*, Chavan A, Freeberg AM, Etwaru D, Ernst DC, Palacios JG, Golden SS, LiWang A, Lander GC, Partch CL
Nat Struct Mol Biol
(bioRxiv https://doi.org/10.1101/2021.9.14.460370)
PDB coordinates for KaiC daytime mimetic – 7S67 (EMD 24852)
PDB coordinates for KaiC nighttime (C6) mimetic – 7S66 (EMD 24851)
PDB coordinates for KaiC nighttime (C2) mimetic – 7S65 (EMD 24850)

Quantification of circadian interactions and protein abundance defines a mechanism for operational stability of the circadian clock
Bagnall JS*, Koch AA*, Smyllie NJ, Begley N, Adamson A, Fribourgh JL, Spiller DG, Meng Q-J, Partch CL, Strimmer K, House, TA, Hastings MH, Loudon ASI
eLife

Cryptochrome proteins regulate the circadian intra-cellular behavior and localization of PER2 in mouse suprachiasmatic nucleus neurons.
Smyllie, N.J., Bagnall, J., Koch, A., Niranjan, D., Poliarova, L., Chesham, J.E., Partch, C.L., Chin, J.W., Loudon, A.S.I., Hastings, M.H. Proc Natl Acad Sci USA

NF-kB modifies the mammalian circadian clock through interaction with the core clock protein BMAL1
Shen Y, Wang W, Endale M, Francey LJ, Harold RH, Hammers DW, Huo Z, Partch CL, Hogenesch JB, Wu Z-H, Liu AC
PLoS Genetics

Reconstitution of an intact clock reveals mechanisms of circadian timekeeping
Chavan AG*, Swan JA*, Heisler J*, Sancar C, Ernst DC, Fang M, Palacios JG, Spangler RK, Bagshaw CR, Tripathi S, Crosby P, Golden SS, Partch CL, LiWang A
Science
(2021) Vol. 374
https://www.science.org/doi/epdf/10.1126/science.abd4453
PDB coordinates for SasA:KaiC CI domain – 6X61
Featured in Rust, M.J. (2021) Biological rhythms: The suspended animation clock. Curr Biol 31: R1513-R1534


Ketogenesis impact on liver metabolism revealed by proteomics of lysine β-hydroxybutyrylation
Koronowski KB, Greco CM, Huang H, Kim, J-K, Fribourgh JL, Crosby P, Partch CL, Qiao F, Zhao Y, Sassone-Corsi P
Cell Reports
(bioRxiv https://doi.org.10.1101/2021.01.21.427645)


CRY2 missense mutations suppress P53 and enhance cell growth
Chan AB, Parico GCG, Fribourgh JL, Ibrahim LH, Bollong MJ, Partch CL, Lamia KA
Proceedings of the National Academy of Science USA
(bioRxiv https://doi.org.10.1101/2021.01.08.425994)

Biochemical mechanisms of period control within the mammalian circadian clock.
Philpott JM, Torgrimson MR, Harold RL and Partch CL
Seminars in Cell and Developmental Biology

The tail of cryptochromes: an intrinsically disordered cog within the mammalian circadian clock
Parico GCG and Partch CL
Cell Communication & Signaling

New insights into non-transcriptional regulation of mammalian core clock proteins.
Crosby P and Partch CL
Journal of Cell Science

Orchestration of circadian timing by macromolecular protein assemblies.
Partch CL
Journal of Molecular Biology
Special issue on Molecular mechanisms underlying circadian regulation
Eds. Eva Wolf and Achim Kramer

The CRY1 tail controls circadian timing by regulating its association with CLOCK:BMAL1.
Parico GCG, Perez I, Fribourgh JL, Hernandez BN, Lee HW, and Partch CL
Proceedings of the National Academy of Science USA
(bioRxiv https://doi.org/10.1011/758714)
Featured in:
-
UCSC Press: Scientists discover how a common mutation leads to a 'night owl' sleep disorder, Tim Stephens
Chemical shift assignments for the human CRY1 tail at BMRB

Dynamics at the serine loop underlie differential ability of cryptochromes for CLOCK:BMAL1 to control circadian timing.
Fribourgh JL*, Srivastava A*, Sandate CR*, Michael AK, Hsu PL, Rakers C, Nguyen LT, Torgrimson MR, Parico GCG, Tripathi S, Zheng N, Lander GC, Hirota T, Tama F, and Partch CL
eLife
(bioRxiv https://doi.org/10.1011/740464)
PDB coordinates for mouse CRY1 PHR:PER2 CBD structure – 6OF7
Coordinates for the PER2 CBD:CRY1 PHR:CLOCK PAS-B HADDOCK model available upon request.

Casein kinase 1 dynamics underlie substrate selectivity and the PER2 circadian phosphoswitch.
Philpott JM*, Narasimamurthy R*, Ricci CG*, Freeberg AM, Hunt SR, Yee LE, Pelofsky RS, Tripathi S, Virshup DM, and Partch CL
eLife
(bioRxiv https://doi.org/10.1011/734624)
Featured in:
-
UCSC Press: Molecular switch mechanism explains how mutations shorten biological clocks, Tim Stephens
-
NIH Director's Blog: Early riser or night owl? New study may help to explain the difference, Francis Collins
PDB coordinates for CK1δ kinase domain structures:
wild-type in anion-free crystallization conditions – 6PXO
tau mutant (R178C) – 6PXN
anion-binding site 2 mutant (R171E) – 6PXP

Regulating behavior with the flip of a translational switch.
Ceh-Pavia E and Partch CL
Proceedings of the National Academy of Science USA
Check out the fantastic paper outlining use of genetic code expansion and noncanonical amino acids to regulate protein expression and circadian behavior from the Hastings and Chin labs here.

CK1d/e protein kinase primes the PER2 circadian phosphoswitch.
Narasimamurthy R, Hunt SR, Lu Y, Fustin J-M, Okamura H, Partch CL,
Forger DB, Kim JK, Virhup DM
Proceedings of the National Academy of Science USA
Chemical shift assignments for mouse PER2 FASP peptide
Check out this great paper from the Okamura lab on a related topic!

Structure, function, and mechanism of the core circadian clock in cyanobacteria.
Swan JA, Golden SS, LiWang A, Partch CL
Journal of Biological Chemistry
Structural dynamics of RbmA governs plasticity of Vibrio cholerae biofilms.
Fong JC, Rogers A, Michael AK, Parsley NC, Cornell WC, Lin YC, Singh PK, Hartmann R, Drescher K, Vinogradov E, Dietrich LE, Partch CL, Yildiz FH
eLIFE
Featured in: Biofilms: Flipping the switch
Pierrat X and Persat A
eLIFE

A slow conformational switch in the BMAL1 transactivation domain modulates circadin rhythms.
Gustafson CL, Parsley NC, Asimgil H, Lee HW, Ahlbach C, Michael AK, Williams OL, Xu H, Davis TL, Liu AC and Partch CL
Molecular Cell
(2017) Vol. 66: 447-457
Featured in: A flick of the tail keeps the circadian clock in line. Narasimamurthy R and Virshup DM
Molecular Cell Vol. 66: 437-438


The assembly and function of bHLH-PAS heterodimers.
Fribourgh JL and Partch CL
Proceedings of the National Academy of Science USA
(2017) Vol. 114: 5330-5332
Commentary on: Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex. Soek SH, Lee W, Jiang L, Molugu K, Li Y, Park S, Bradfield CA, Xing Y.
Proceedings of the National Academy of Science USA
(2017) Vol. 114: 5431-5436

Structural basis of the day-night transition in a bacterial circadian clock.
Tseng R*, Goularte NF*, Chavan A*, Luu J, Cohen SE, Chang YG, Heisler J, Michael AK, Tripathi S, Golden SS, LiWang A, Partch CL
Science
(2017) Vol. 355: 1174-80
PDB coordinates for Kai complex structures:
fold-switch KaiB:KaiC CI domain – 5JWO
fold-switch KaiB:KaiC S431E hexamer (left) – 5JWQ
KaiA deltaN:fold-switch KaiB:KaiC CI domain – 5JWR
fold-switch KaiB:CikA PsR domain – 5JY5

Formation of a repressive complex in the mammalian circadian clock is mediated by the secondary pocket of CRY1.
Michael A, Fribourgh J, Chelliah Y, Sandate C, Hura G, Schneidman-Duhovny D, Tripathi SM, Takahashi JS, Partch CL
Proceedings of the National Academy of Science USA
(2017) Vol. 114: 1560-65

PDB coordinates for mouse CRY1 PHR structure – 5T5X
Coordinates for HADDOCK, MultiFoXS, and FoXSDocK models available upon request.

Animal Cryptochromes: Divergent roles in light perception, circadian timekeeping and beyond.
Michael A*, Fribourgh J*, Van Gelder RN, Partch CL
Photochemistry and Photobiology
(2017) Vol. 93: 128-40
Special issue in honor of Aziz Sancar, 2015 Nobel Laureate in Chemistry


Early doors (Edo) mutant mouse reveals the importance of period 2 (PER2) PAS domain structure for circadian pacemaking.
Militi S*, Maywood ES*, Sandate CR, Chesham JE, Barnard AR, Parsons MJ, Vibert JL, Joynson GM,Partch CL, Hastings MH, Nolan PM
Proceedings of the National Academy of Science USA
(2016) Vol. 113: 2756-61
SAXS profiles for WT & Edo PER2 PAS-AB domains on BioISIS.net

Cytosolic BMAL1 moonlights as a translation factor.
Michael AK, Asimgil H, Partch CL
Trends in Biochemical Science
(2015) Vol. 40: 489-90

Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C-terminus
Xu H*, Gustafson CL*, Sammons PJ, Khan SK, Parsley NC, Ramanathan C, Lee HW, Liu AC, Partch CL
Nature Structural and Molecular Biology
(2015) Vol. 22: 476-84
Featured in: Grab the wiggly tail: new insights into the dynamics of circadian clocks. Hui KY and Ripperger JA
Nature Structural and Molecular Biology (2015) Vol. 22: 435-36
Chemical shift assignments for BMAL1 and BMAL2 TADs

Cancer/Testis antigen PASD1 silences the circadian clock.
Michael AK, Harvey SL, Sammons PJ, Anderson AP, Kopalle HM, Banham AH, Partch CL
Molecular Cell
(2015) Vol. 58: 743-54
Featured in:
-
UCSC Press: Scientists discover protein that silences biological clock, Tim Stephens
-
Quanta Magazine: How the body's trillions of clocks keep time, Veronique Greenwood

Analysis of protein stability and ligand interactions by thermal shift assay.
Huynh K and Partch CL
Current Protocols in Protein Science
(2015) Vol. 79: 28.9.1-14
Additional information on acquiring and processing thermal shift data available on Resources page

Coiled-coil coactivators play a structural role mediating interactions in hypoxia-inducible factor heterodimerization.
Guo Y, Scheuermann TH, Partch CL, Tomchick DR, Gardner KH
Journal of Biological Chemistry
(2015) Vol. 290: 7707-21

Emerging models for the molecular basis of mammalian circadian timing.
Gustafson CL and Partch CL
Biochemistry
(2015) Vol. 54: 134-49

Antibacterial membrane attack by a pore-forming intestinal C-type lectin.
Mukherjee S, Zheng H, Derebe MG, Callenberg KM, Partch CL, Rollins D, Propheter DC, Rizo J, Grabe M, Jiang QX, Hooper LV
Nature
(2014) Vol. 505: 103-7

Molecular architecture of the mammalian circadian clock.
Partch CL, Green CB, Takahashi JS
Trends in Cell Biology
(2014) Vol. 24: 90-9



